In the sphere model all proteins pretty much look alike. But I still think kind of cool, especially if you use Lightnir's QuteMol preset for pymol.
 The cartoon model shows you more about the structure. GFP consists of a β-sheet "barrel" that encapsulates the chromophore. You need a rigid structure if you want to make sure that the chromophore does not quench before it is able to show fluorescence. It is interesting to compare this to Rhodopsin which is apparently much more flexible. This makes sense since in Rhodopsin the biological activity is related to non-radiative decay and isomerisation.
The cartoon model shows you more about the structure. GFP consists of a β-sheet "barrel" that encapsulates the chromophore. You need a rigid structure if you want to make sure that the chromophore does not quench before it is able to show fluorescence. It is interesting to compare this to Rhodopsin which is apparently much more flexible. This makes sense since in Rhodopsin the biological activity is related to non-radiative decay and isomerisation. The chromophore is trapped at the center of the barrel.
The chromophore is trapped at the center of the barrel.
Interestingly the chromophore is made directly out of amino acids. You can take a look at it and think of how this is done.
 The answer is here. The hydroxyphenyl comes from tyrosine (as expected). The imidazolon ring is formed after cyclization, the bridging double bond through oxidation. It is interesting to consider that this apparently works in many different organisms and not just in the jelly fish aequoria victoria. The driving force seems to be that the amino acids are pressed together in the barrel.
The answer is here. The hydroxyphenyl comes from tyrosine (as expected). The imidazolon ring is formed after cyclization, the bridging double bond through oxidation. It is interesting to consider that this apparently works in many different organisms and not just in the jelly fish aequoria victoria. The driving force seems to be that the amino acids are pressed together in the barrel.The chromophore is part of the protein chain. It is connected to the helix that goes through the center of the protein.
 Or from a different perspective.
Or from a different perspective.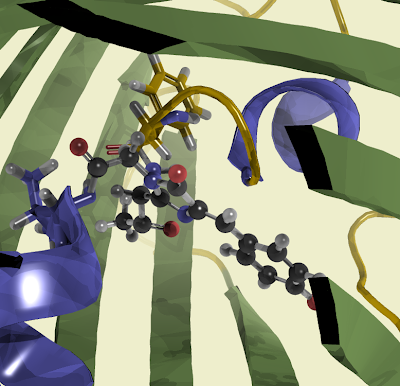
9 comments:
Every time I come here I get amazed in how many ways the same molecule can be presented. The 4th and last render of GFP are really gorgeous. The chromophore looks like it was made of some sort of shiny plastic or glass. Did you tweak the light source settings (ambient/specular) or used the default light values and played only with the representations?
thanks. it's kind of fun to play around with these things. actually that shininess just comes from those QuteMol settings. what i changed is just perspective, colors, and cut-off. the color for turns/loops was gold from here: http://pldserver1.biochem.queensu.ca/~rlc/work/pymol/
maybe that's also why it shines ...
Hi Felix,
Your pictures of molecules are beautiful. Could you look at my molecular graphics? Maybe you may advise how to improve it?
http://www.agilemolecule.com/Products.html
Best wishes,
Alexey
hi, it's definitely a nice editor with pretty representations
the nano-structures editor is really nice.
did you include a way to export pictures, some raytracing?
Thank you. The raytracing is under discussion. I think that export will be done at the final stage when graphics become stable. It's seems to me that its quality far from desirable.
i think it has a nice feel, but of course competion is hard in this sector ...
can i put up some pictures of nano-structures? or is licensing needed even with private just for fun use?
You can take pictures. But Ascalaph has no nano editor. This is only examples. They are made using Nanotube Modeler from www.jcrystal.com and "Add" command of Ascalaph.
thanks. i think those templates in the AscalaphDesignerDemo are already pretty nice to play around with a little bit
Demo does not allow to save files. AscalaphQuantum has the same features. Hope you enjoy.
Post a Comment